PCam: histopathology dataset for fundamental machine learning.
During my work[1] on deep learning models for histopathology, I’ve started to appreciate the tremendous barrier-to-entry that exists for machine learning researchers to evaluate their methods on large medical datasets. This is especially the case for histopathology, where large datasets such as Camelyon require developing intricate dataloaders, that take care of balancing different types of tissue, and can efficiently work with the terabytes of data available. With the hopes that we will make life a little easier for researchers looking to work with histopathology data, we are launching a dataset derived from Camelyon: PatchCamelyon (PCam). It was developed with input from pathologists and experts on deep learning for histopathology.
PCam is available now at github.com/basveeling/pcam, and is just as straight forward to work with as CIFAR10 or MNIST.
Excerpt from the Readme:
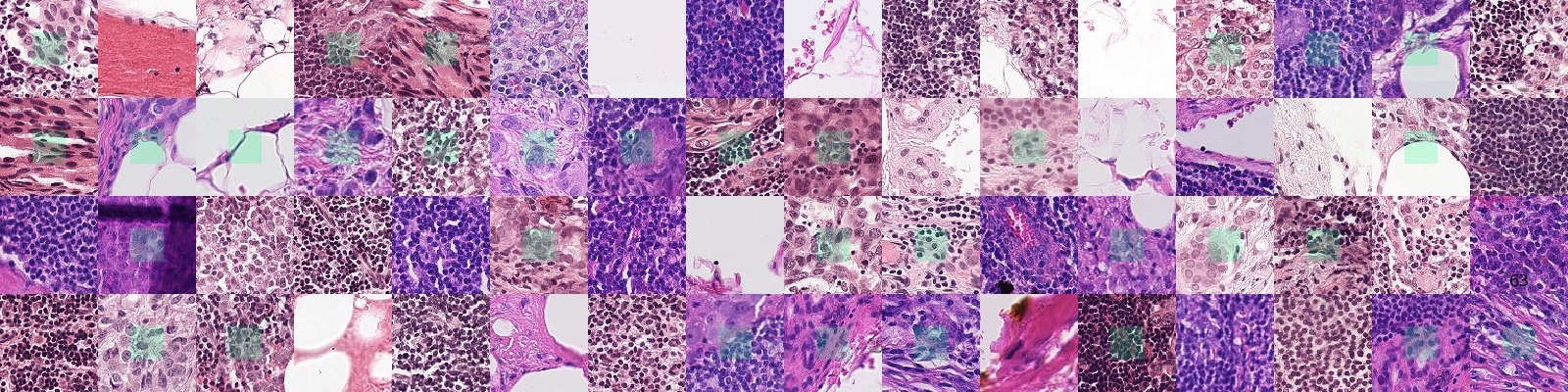
The PatchCamelyon benchmark is a new and challenging image classification dataset. It consists of 327.680 color images (96 x 96px) extracted from histopathologic scans of lymph node sections. Each image is annoted with a binary label indicating presence of metastatic tissue. PCam provides a new benchmark for machine learning models: bigger than CIFAR10, smaller than imagenet, trainable on a single GPU.
Why PCam
Fundamental machine learning advancements are predominantly evaluated on straight-forward natural-image classification datasets. Think MNIST, CIFAR, SVHN. Medical imaging is becoming one of the major applications of ML and we believe it deserves a spot on the list of go-to ML datasets. Both to challenge future work, and to steer developments into directions that are beneficial for this domain.
We think PCam can play a role in this. It packs the clinically-relevant task of metastasis detection into a straight-forward binary image classification task, akin to CIFAR-10 and MNIST. Models can easily be trained on a single GPU in a couple hours, and achieve competitive scores in the Camelyon16 tasks of tumor detection and WSI diagnosis. Furthermore, the balance between task-difficulty and tractability makes it a prime suspect for fundamental machine learning research on topics as active learning, model uncertainty and explainability.
- [1] B. S. Veeling, J. Linmans, J. Winkens, T. Cohen, M. Welling, Rotation Equivariant CNNs for Digital Pathology. arXiv [cs.CV] (2018), (available at http://arxiv.org/abs/1806.03962).

Leave a Comment
Your email address will not be published. Required fields are marked *